Job Objects: The Keys to Data and Results
Introduction
In Accelerator terms, a job is an executed program, it contains everything input to as well as everything generated by a program run, and it is stored on disk. The job object is used to represent a job, and it is a keeper of links to datasets, files, parameters, and metadata.
The job object is used to pass data and files between jobs, thereby creating dependencies, and between jobs and build scripts. Job objects are easy to use, and bring a set of functions that makes common tasks straightforward to write and comprehend. This post gives an overview of most of the job object’s possibilities.
Job Objects are Returned from Build Calls
The first place we encounter job objects is in build scripts. The Accelerator uses build scripts to execute jobs. Jobs are stored on disk, so that they can be used by other jobs and re-used if the same build call is run again. Here is an example of a simple build script:
def main(urd):
job = urd.build('csvimport', filename='mydata.csv')
This build script will create a job containing everything related to
the execution of the method csvimport with the option
filename='mydata.csv. When execution finishes, the call returns a
job object representing the job.
Job Objects Represent Either Existing Jobs or the Currently Running Job
To be specific, job objects come in two very similar flavours. There is one used to reference existing jobs, for example in build scripts as shown in the example above, and one used inside running jobs, used to represent the running job itself. The vast majority of functionality is available in both places, but functions that only makes sense in one of the places is unavailable in the other. The difference is kind of obvious when working on a project, so it will not be mentioned again here.
1. Job References
The most common use of the job object in a build script is to make the job available to new jobs, like in this example:
job_import = urd.build('csvimport', filename='mydata.csv')
job_analyse = urd.build('analyse', source=job_import)
Here, job_analyse has access to everything related to job_import,
since it is using job_import as an input parameter.
2. Datasets
The Accelerator’s Dataset is a data structure for tabular data, that is designed for efficient parallel processing.
There is a tight connection between jobs and datasets. Datasets are created by, and stored in, jobs.
Are There any Datasets in the Job?
This will return a list of all datasets in a job:
all_datasets = job.datasets
How do I get a Reference to a Named Dataset?
To get a reference to a particular named dataset, use the job.dataset() function:
ds = job.dataset(name)
An empty name corresponds to the default dataset, i.e. name='default'.
How do I Create a Dataset in a Job?
Datasets are created using job.datasetwriter(). Creating datasets
is out of this posts’ scope, but here’s a sketch:
def prepare(job):
dw = job.datasetwriter(name='movies')
dw.add('moviename', 'unicode')
return dw
def analysis(sliceno, prepare_res):
dw = prepare_res:
...
dw.write(data)
See the manual or other examples or posts for more information.
3. Files
There are three ways for a job to store data: as datasets, files, or using the return value, where the return value is actually a special case of a file.
How to Load and Store Data in Files
To simplify the coding experience, the Accelerator has built in support for storing and retrieving data in Python’s pickle format or in JSON format.
A running job can use job.save() to store data to a file in a job.
The data will be serialised using Python’s pickle format. For
example:
def synthesis(job):
data = ...
job.save(data) # using default name "result.pickle"
job.save(data, 'anothername.pickle')
return data # store data as "result.pickle"
Note the last line - the data returned from synthesis is automatically
stored in the file result.pickle!
Loading data from a job is equally simple, using job.load(), like this:
data = job.load('anothername.pickle')
data = job.load() # to load "result.pickle"
A common example would be to run a job and then present its output
somehow. Assume that the analyse job returns some data using the
return statement, then we could do:
job_analyse = urd.build('analyse', source=job_import)
data = job_analyse.load()
# do something with data here, for example
print(data)
The power of using the pickle format is that it is possible to store and retrieve complex datatypes between jobs and build scripts without having to consider formatting or parsing!
The JSON-equivalents are called job.json_save() and
job.json_load(). (The Accelerator typically uses JSON format
internally for meta data files and pickle format for everything else.)
It should be mentioned that job.open() as well as the load and save
functions can work on parallel files, i.e. files that are written
in parallel independently in each slice/process by one job, and read back in
parallel in another job. See manual for more information!
How can I Find the Names of all Files Stored in a Job?
all_files = job.files()
This will return a set of all files created by the job. This
function takes an optional pattern argument that is used to filter
the filenames:
only_the_text_files = job.files('*.txt')
So, How does the Job Know what Files it Contains?
The function job.save(), as well as its JSON counterpart, will
register its files automatically to the running job. If files are
generated by some other means, files can be registered manually, like
this:
def synthesis(job):
...
PIL.Image.save(filename) # saves a file using Python Image Library
job.register_file(filename)
or, using the extended job.open(), a wrapper for plain open() that
will register the file automatically:
def synthesis(job):
...
with job.open(filename, 'wb') as fh:
fh.write(mydata)
How to get the absolute filename to a file in a job?
If the name of the file is known,
absname = job.filename(name)
will return the full pathname to the file.
4. Transparency
There are several ways to inspect a job.
-
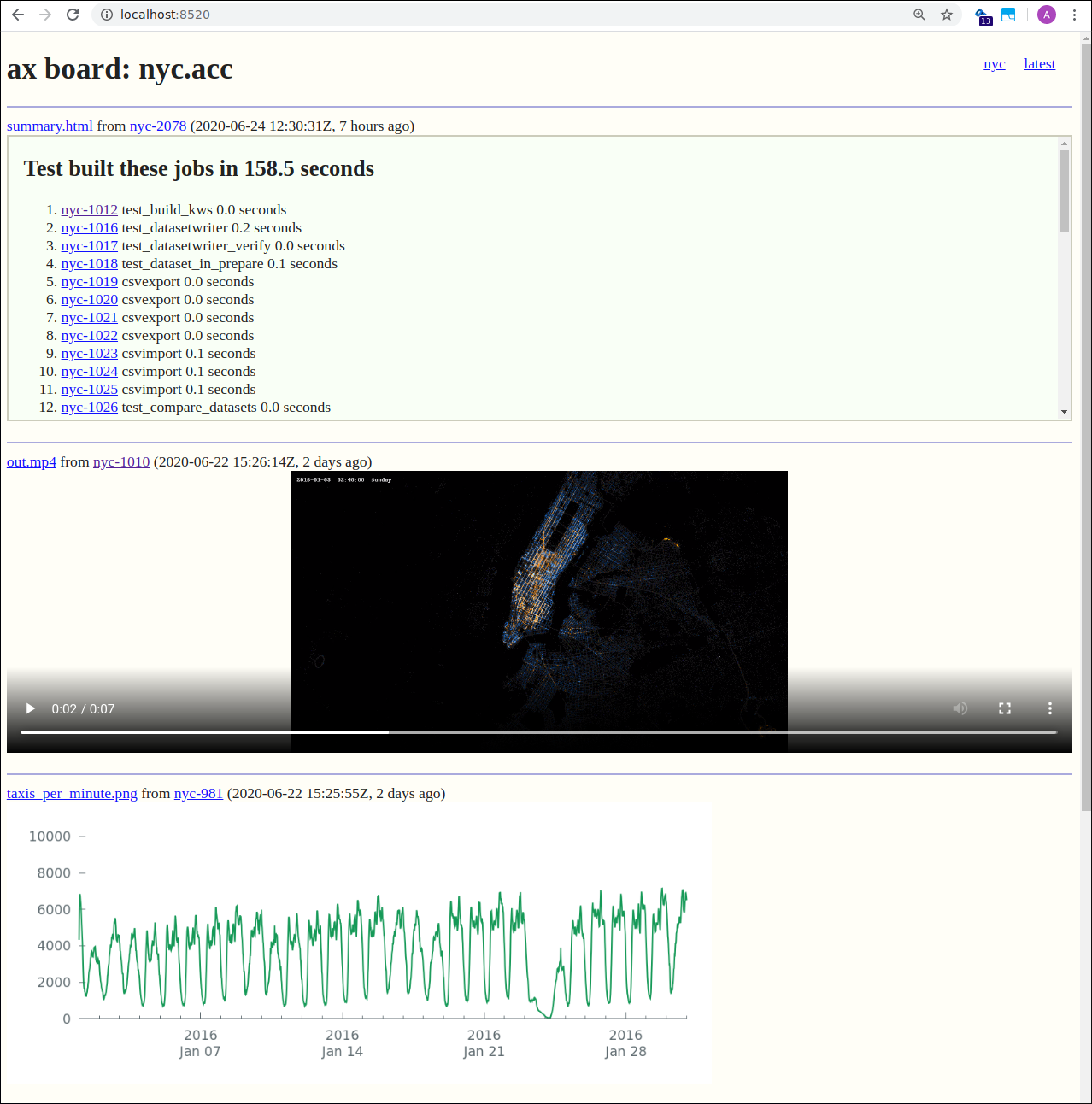
Using the Accelerator Board web server and inspect jobs and datasets from your web browser. Launch it by typing
ax board-serverand connect to it using a web browser pointing to
http://localhost:8520(Edit
accelerator.confto have it autostarting listening to any port of choice.) -
Use the
ax job <job>command to get information on a specific job -
All job meta-data that makes sense to a human is stored in JSON-format, so it is possible to navigate to a job directory and inspect the files directly, in particular, the files
setup.jsonandpost.json.
Here is an example using the board server
5. Results
The result directory is a place where “global” results and findings can be put. We use soft links, that point to files in jobs, instead of putting actual files there. The reason is that if a files is copied from a job to the result directory, the connection between the file and its origin (the job) is gone. The difference between a file and a link is that the link shows the actual location of the original file (which is in the job that created it). So the link maintains transparency.
Making a File in a Job Visible in the Result Directory
The preferred and easiest way to create links in the result directory
is to use job.link_result(), like this:
job = urd.build('plotsomething', source=mydataset)
job.link_results('plot.png')
This will create a soft link in the result directory pointing to the
file plot.png in job.
The name of the link can be set using the linkname option:
job = urd.build('plotsomething', source=mydataset)
job.link_results('plot.png', linkname='distribution_of_taxicabs_on_mondays.png')
where linkname= is optional.
6. File references
Passing jobs as input to other jobs is, as we’ve seen, straightforward. But sometimes we only want to pass specific files from one job to another. There are several ways to do this:
1. Using the job object only
We pass the job object representing an existing job with files to a new job. In this case, the new job is thus informed about the job that created and holds the files, but it will not know the name of the actual files.
If there is only one file, and we use the convention to name the file
result.pickle, we can load the contents of the file using
job.load(). But if there are more than one file, or the file name
is not always unique, the new job does not know how to resolve the
situation. One solution is to hard code the file names in the new
job’s source code. This is ok in many cases, but it is not a generic
solution.
2. Pass the filename as parameter (but please don’t)
We pass the full pathname of the file as a parameter to the new job. In this case, the new job can read the file directly, but the information about which job that generated the file is gone. This breaks transparency and should be avoided.
3. Pass the file name together with a reference to the job
The job.withfile() function is used for that, like this:
job = urd.build('create_two_files')
urd.build('checkfile', thefile = job.withfile('fileA')
urd.build('checkfile', thefile = job.withfile('fileB')
Here, the create_two_files job (implicitly) creates the files
fileA and fileB. The checkfile method is then built twice,
once for each of the filenames in the “source” job job.
Inside the method (in this case checkfile), it looks something
like this
from accelerator import JobWithFile
options = dict(thefile=JobWithFile())
def synthesis():
filename = options.thefile.filename()
#
data = options.thefile.load()
# or
data = options.thefile.load_json()
# or
with options.thefile.open('rb') as fh:
data = fh.read()
7. Miscellaneous Functions
Miscellaneous Functions, Available for Existing Jobs
These functions are available on existing jobs to investigate a job’s output, input parameters, and execution times (covering both parallel and serial parts of the program in detail).
job.output() # A string containing everything the job has written to stdout and stderr!
job.params # A dict containing all the job's parameters
job.post # Execution times for all parts of the job
Miscellaneous Functions, Available for Running Jobs
Information on location of input and result directory is available to the running job object:
job.input_directory
The reference to the input_directory is useful when writing data
import methods. Storing data in the input_directory, specified by
the Accelerator’s configuration file, makes it possible to import data
without absolute file path dependencies. This makes it much easier to
port a project to a different system.
Miscellaneous Functions, Common for both Existing and Running Jobs
These are only mentioned for completeness:
job.method # name of the job's method (i.e. corresponding source filename)
job.path # absolute path of job directory
job.workdir # name of workdir where the job is stored
job.number # the integer part of the jobid.
8. Job Chains
Similar to dataset chains, it is possible to create chains of jobs. A
job chain is created using the jobs.previous input parameter. In a
method:
jobs = ('previous',)
def synthesis():
...
and in the build script
previous = None
for ix in range(10):
previous = urd.build('thejob', previous=previous)
This creates a job chain that is ten jobs long. The
job.chain()-function will return a list of all jobs in the chain.
list_of_jobs_in_chain = job.chain()
Similar to dataset chains, this function takes some options, see documentation for details.
9. Relation to Other Classes
The graph below shows as simplified version of how the different classes used by the Accelerator are connected. The classes at the top are used in build scripts, and classes at the bottom in running jobs.
Note that
-
only a small subset of all available class member functions are shown in order to keep the image simple, and
-
the graph does only show the most relevant classes.
10. Conclusion
The intention of this post is to show the various functions and most common use cases for the job object. The job object provides streamlined solutions to common situations that reduce the amount of code to write (and maintain). For more details and practical examples, see the references below.
Additional Resources
The Accelerator’s Homepage (exax.org)
The Accelerator on Github/exaxorg
The Accelerator on PyPI
Reference Manual